Appearance
Map Viewer
Core Feature
This feature is available in all MATISSE Explorer deployments.
The Map Viewer is the primary visualization component in MATISSE Explorer. It displays your spatial transcriptomics data as an interactive map, allowing you to explore tissue images overlaid with data points.
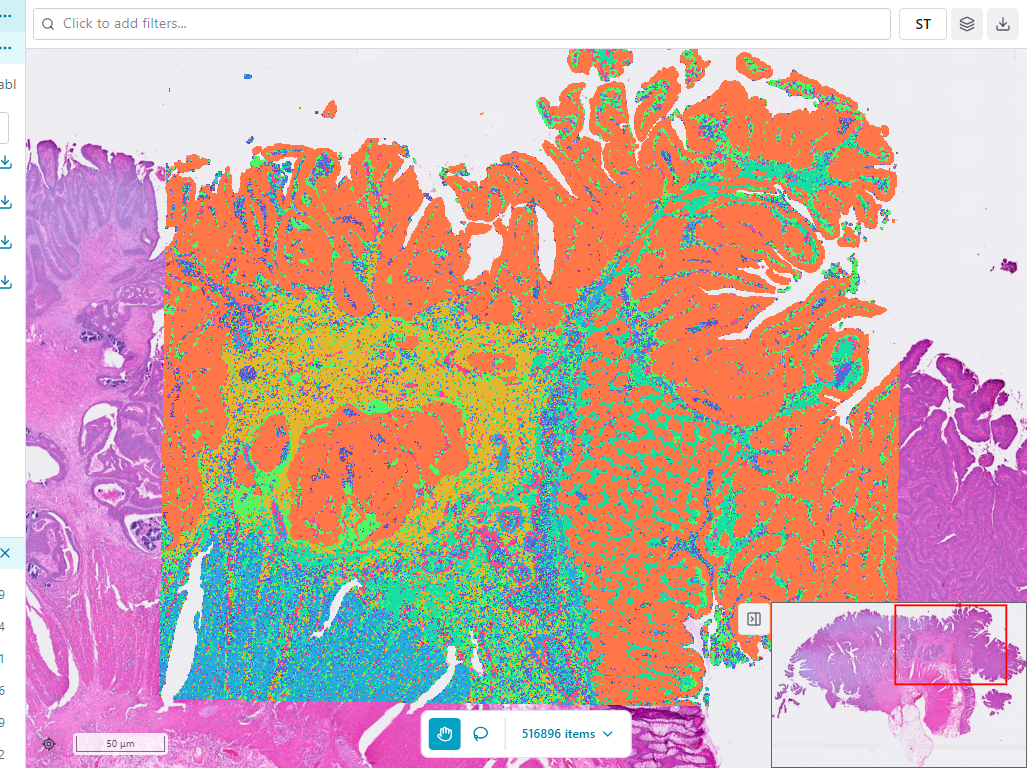
Interface Components
Toolbar
The toolbar at the top of the map viewer provides access to essential controls:

| Control | Description |
|---|---|
| Pan (Hand icon) | Navigate the map by clicking and dragging |
| Selection (Lasso icon) | Draw selections to highlight regions of interest |
| Items dropdown | Shows count of visible/subset items with additional options |
Map Area
The central map area displays:
- Background image - Tissue image (if available)
- Data points - Individual cells or spots colored by the selected feature
- Selection overlay - Highlighted region when using lasso selection
Legend
The legend shows the color mapping for the current feature:
- Categorical features - List of categories with their colors
- Continuous features - Color gradient with min/max values
Hover over legend items to highlight corresponding points on the map.
Subset Panel
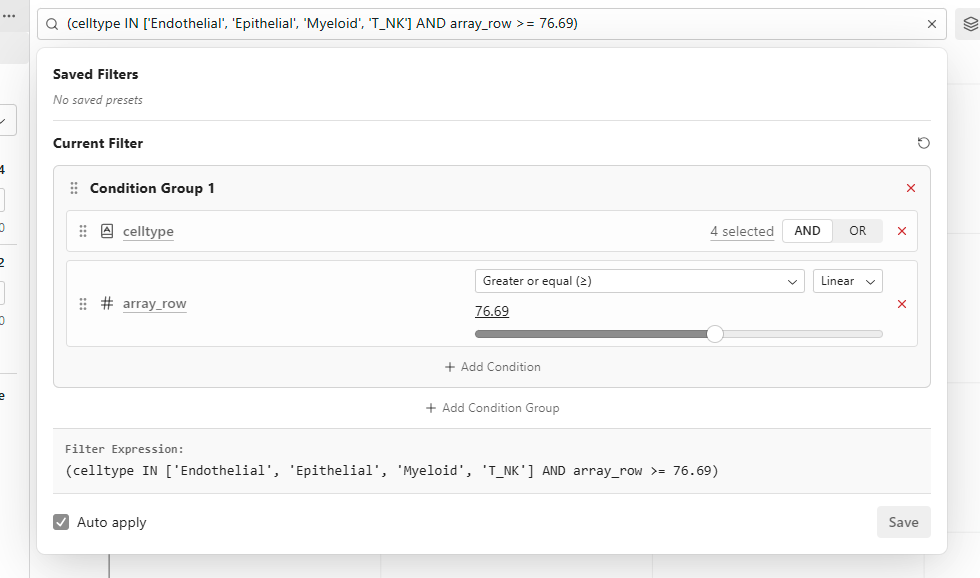
Click the subset input field to open the Subset Panel. See Subset for details.
Interaction Modes
Pan Mode
Pan mode is the default interaction mode for navigating the map.
Controls:
| Action | Mouse | Result |
|---|---|---|
| Pan | Click + drag | Move the map view |
| Zoom in | Scroll up / Double-click | Zoom into the map |
| Zoom out | Scroll down | Zoom out of the map |
Selection Mode
Selection mode allows you to draw lasso selections to highlight specific regions.
How to use:
- Click the Selection button (Lasso icon) in the toolbar
- Click and drag on the map to draw a closed shape
- Release to complete the selection
- The toolbar shows the count of selected items
After selecting:
Click the Items dropdown to access options:
- Add selection to subset - Save the lasso as a subset condition
Projection Modes
Switch between different coordinate systems to view your data from multiple perspectives.
Available Projections
| Mode | Description |
|---|---|
| Spatial | Original tissue coordinates (default) |
| UMAP | UMAP dimensionality reduction coordinates |
| t-SNE | t-SNE dimensionality reduction coordinates |
Switching Projections
- Locate the projection selector in the toolbar or sidebar
- Click to open the dropdown menu
- Select the desired projection mode
- The map view updates to show the new coordinates
Lasso selections made in embedding space (UMAP/t-SNE) can be saved and reused in subsets.
Supported Image Formats
The Map Viewer supports multiple image formats for background tissue images:
| Format | Description | Best For |
|---|---|---|
| GeoTIFF | Cloud Optimized GeoTIFF | Large images, streaming |
| Static | PNG, JPEG | Small to medium images |
| Tiles | Multi-resolution tile pyramid | Very large images |
The appropriate format is determined by your data configuration and loaded automatically.
Tooltip
Hover over data points to see detailed information:
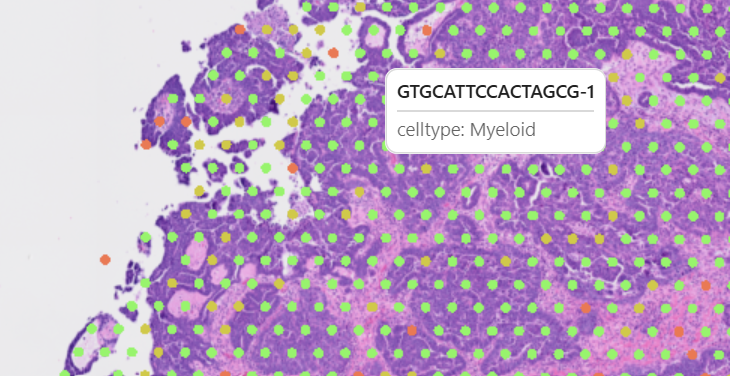
The tooltip displays:
- Current feature value for the hovered point
- Point index or identifier
- Additional metadata (if configured)
Performance Tips
Large Datasets
For datasets with millions of points:
- Use tile-based images when available
- Allow time for initial rendering
- Zoom into regions of interest for smoother interaction
- Consider using subsets to reduce visible points
Smooth Navigation
- Avoid rapid zoom changes
- Wait for rendering to complete before making new selections
- Use Pan mode for general navigation, switch to Selection only when needed
Related Topics
- Navigation & Zoom - Detailed navigation controls
- Lasso Selection - Complete lasso selection guide
- Color Mapping - Configure data point colors
- Layer Settings - Adjust point size and opacity