Appearance
Quick Start Guide
Core Feature
This feature is available in all MATISSE Explorer deployments.
Get started with MATISSE Explorer in just a few minutes. This guide walks you through the essential steps to explore your spatial transcriptomics data.
Step 1: Open Your Project
InSilicoLab Users
If you are using InSilicoLab, select your project from the Reports section to open MATISSE Explorer.
When you launch MATISSE Explorer, your project data loads automatically. You will see:
- The map viewer displaying your tissue image with data points
- The Sidebar showing available features
- Loading indicators while data is being fetched
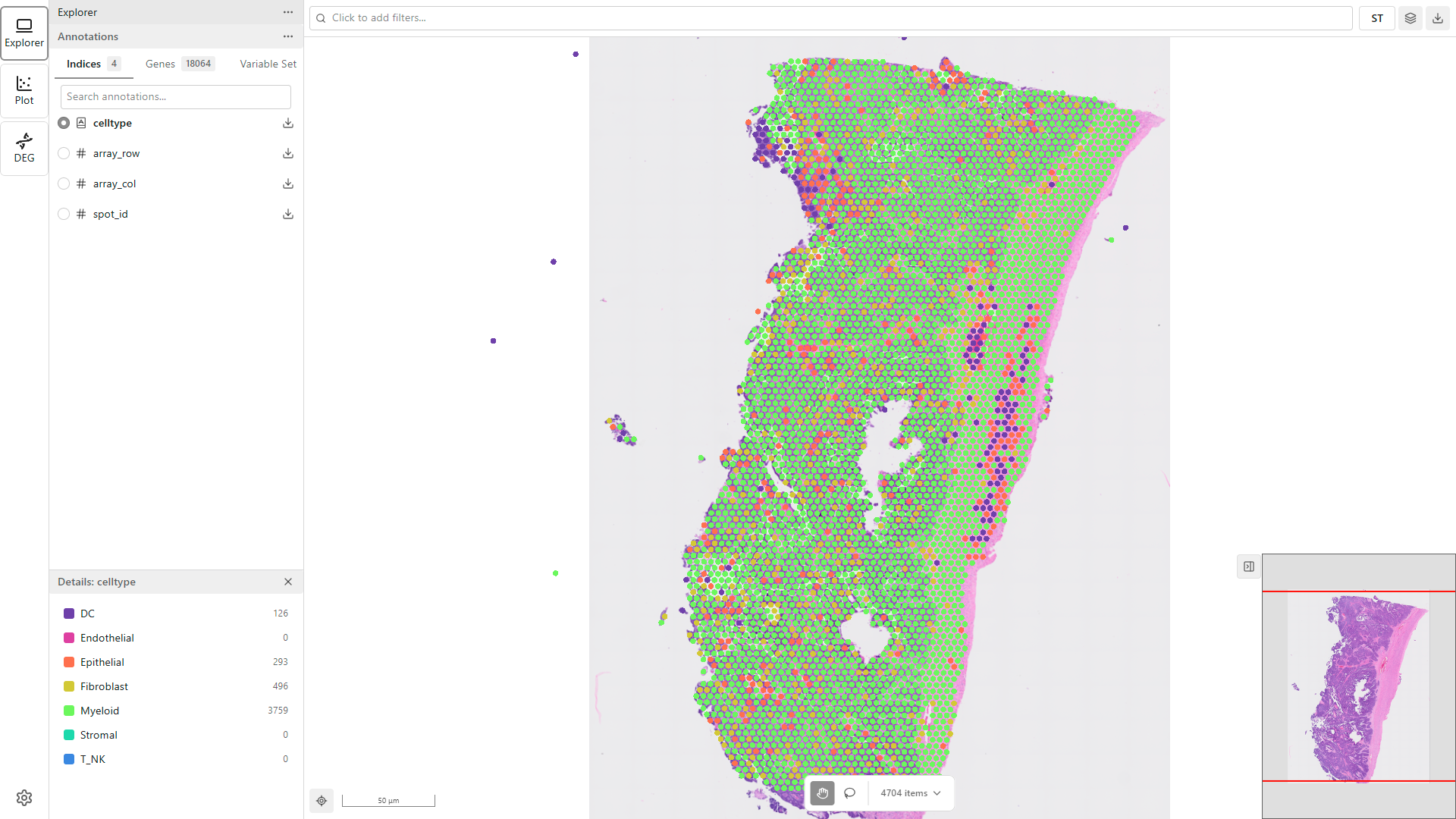
Wait for the data to finish loading before proceeding. Large datasets may take a moment to fully render.
Step 2: Color by Feature
Apply color mapping to visualize patterns in your data:
- In the Sidebar, browse the Features list
- Click the Categorical tab to see cell type features
- Click on a feature name (e.g., "cell_type") to select it
- The map updates to show data points colored by the selected feature
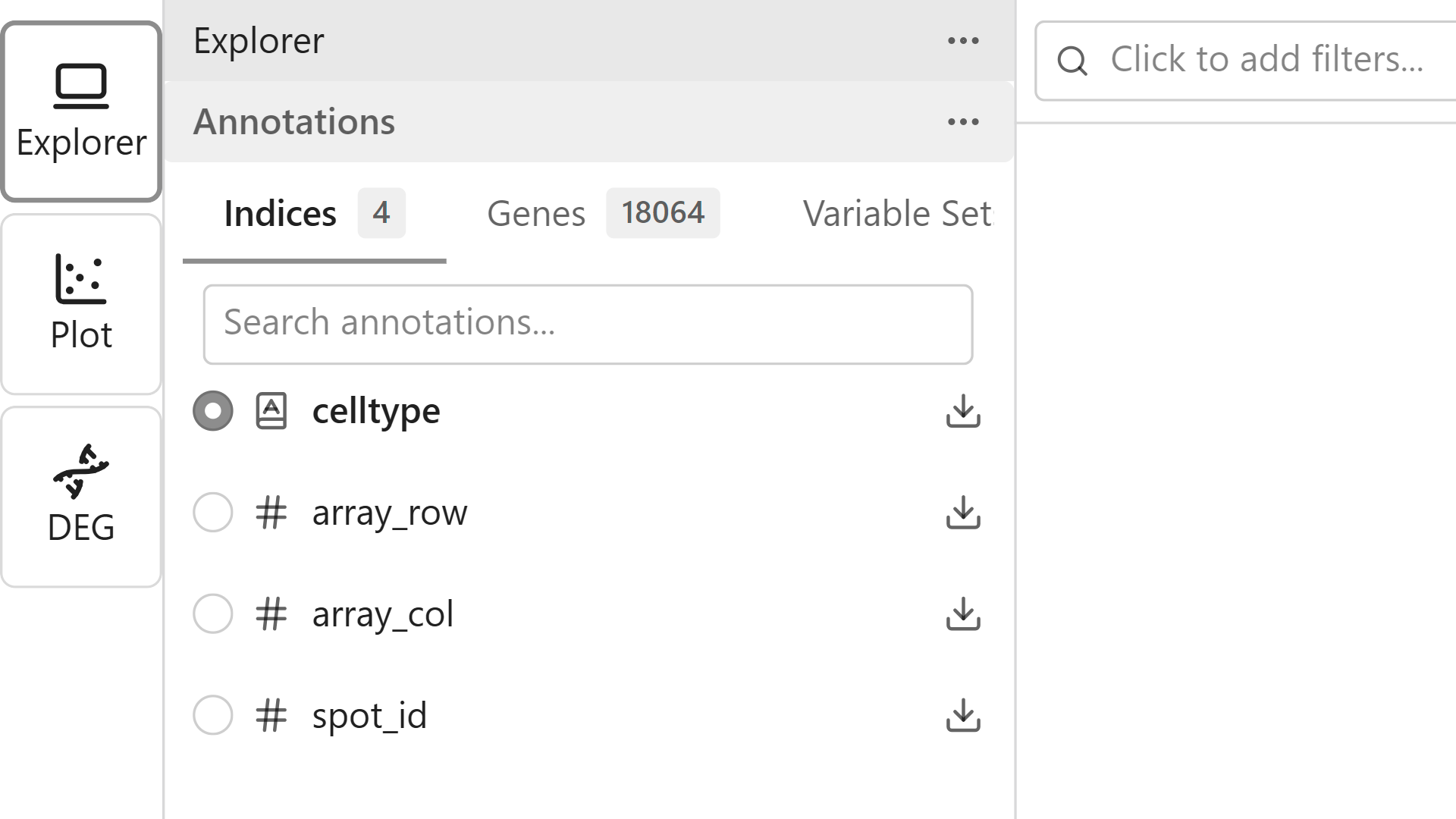
Try Different Feature Types
- Categorical - Discrete categories like cell types or clusters
- Continuous - Numeric values like gene expression levels
- Embeddings - Dimensionality reduction coordinates
Step 3: Navigate the Map
Explore your data using mouse controls:
| Action | Control |
|---|---|
| Pan | Click and drag |
| Zoom in | Scroll wheel up or double-click |
| Zoom out | Scroll wheel down |
| Reset view | Click the reset/home button in toolbar |
Step 4: Apply a Subset
Narrow down your data to specific populations:
- Click the subset input field in the toolbar to open the Subset Panel
- Click Add Condition to create a new subset rule
- Select a feature and set your criteria
- The subset applies automatically (Auto apply is enabled by default)
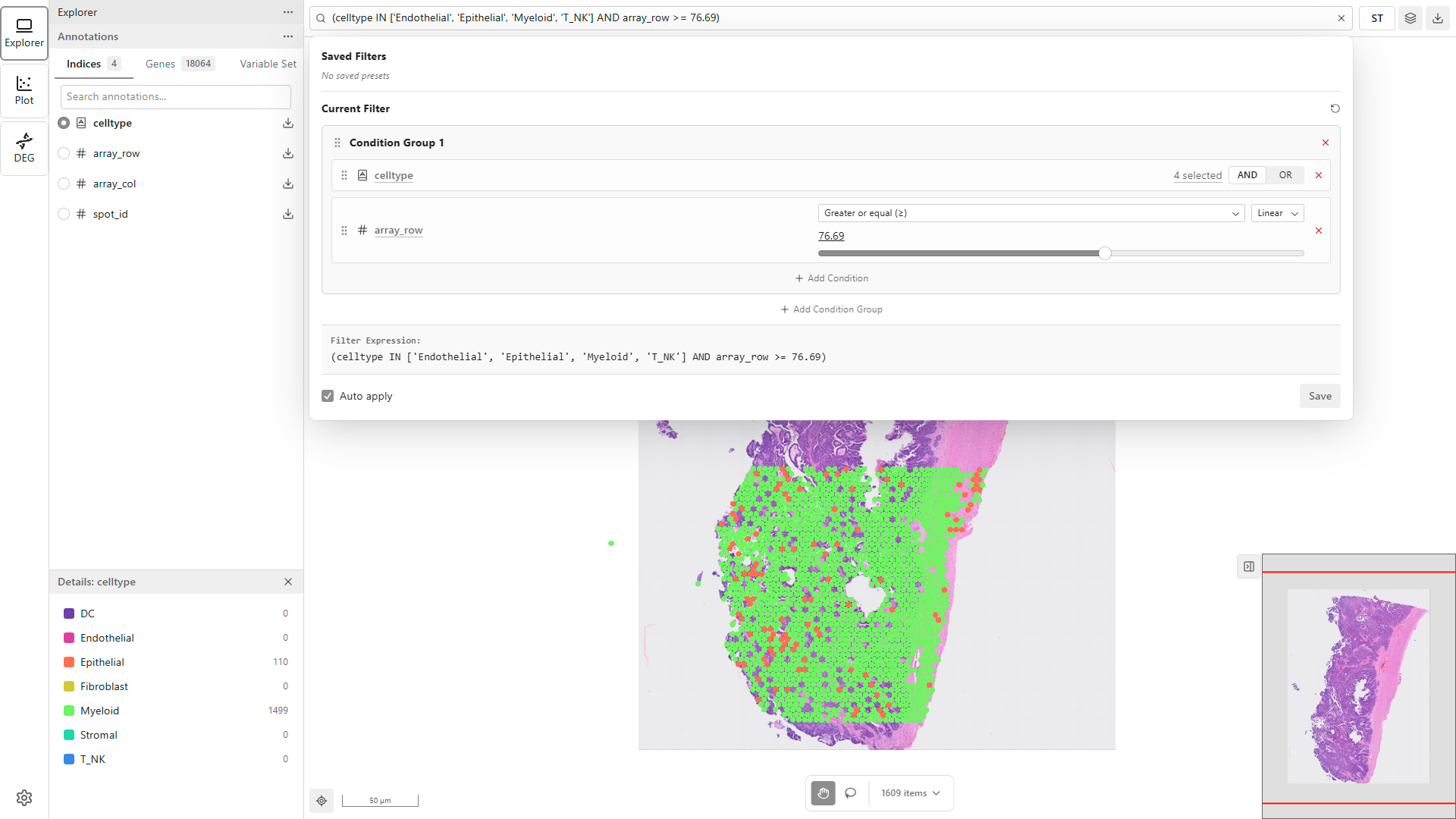
Only data points matching your subset criteria will remain visible. The point count updates to show how many points pass the subset.
Step 5: Use Lasso Selection
Select specific regions interactively:
- Click the Selection mode button in the toolbar
- Draw a closed shape around your region of interest
- Release to complete the selection
- View the count of selected points
What You Can Do With Selections
- Add the selection as a subset condition
- View statistics for selected points
- Export selected data
Step 6: View a Scatter Plot
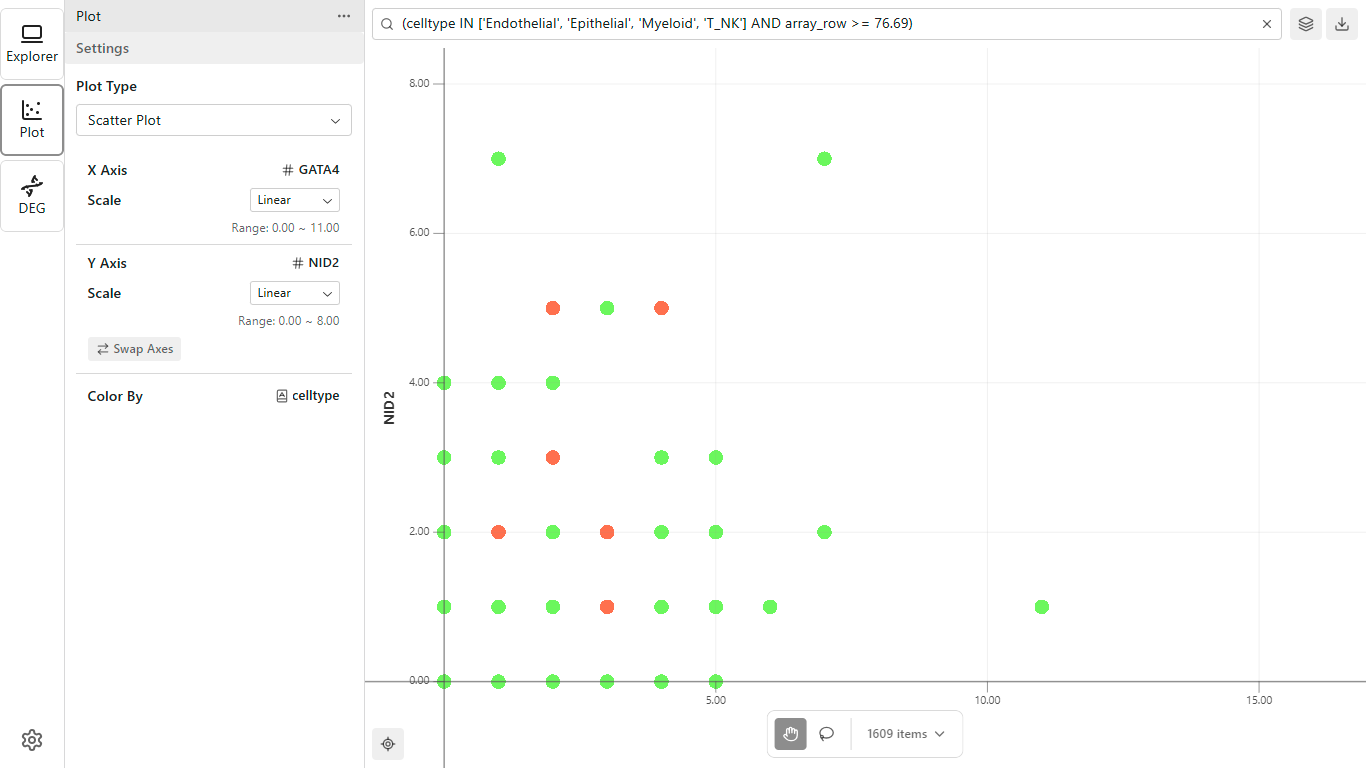
Explore relationships between features:
- Click the Plot icon in the Activity Bar
- In the Sidebar, configure your X and Y axes
- Optionally set a color mapping
- The scatter plot renders in the Workspace
Next Steps
Now that you know the basics, explore more features:
- Map Viewer - Detailed map viewer documentation
- Subset - Build complex subset queries
- Signatures - Create custom gene groups
- Tutorials - Step-by-step workflow guides
Tips for New Users
- Start with categorical features - They provide clear visual groupings
- Use the legend - Hover over legend items to understand the color mapping
- Save subsets - Reuse complex subset configurations
- Try different projections - Switch between spatial, UMAP, and t-SNE views
- Export early, export often - Save subset data for downstream analysis